Tags
anatomically modern humans, David Reich, Denisovans, genetic distance, genus, Neanderthals, race, species, taxonomy
About a year ago I wrote:
The following chart (created by some scientist(s) led by David Reich) shows the genetic divergence between hominin samples as a fraction of the human-chimp difference. So for example, all the human groups have just over a 0.12 genetic divergence with Neanderthals, meaning that the genetic difference between humans and Neanderthals is only 12% as great as the genetic difference between humans and Chimps (source: supplement of Genetic history of an archaic hominin group from Denisova Cave in Siberia.)

The purpose of the chart is to estimate how long ago the different populations diverged from a common ancestor. So since the fossil record tells us that Neanderthals and chimps diverged about 6.5 million years, then humans and Neanderthals should have diverged roughly 0.8 million years ago (12% of 6.5 million) assuming genetic divergence maps to chronological divergence in a linear way.
I transformed the genetic distance matrix into a dendrogram, which looks at all the distances and creates the most parsimonious family tree:
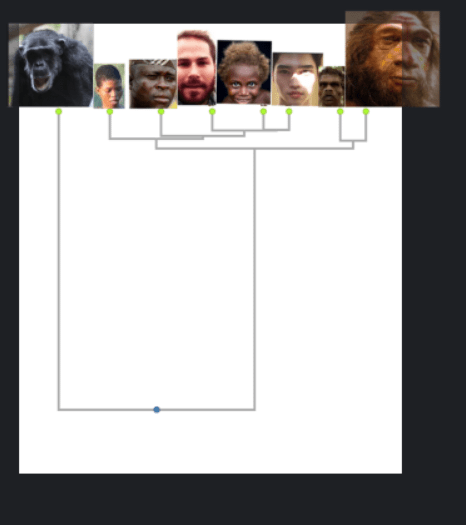
What’s cool about dendrograms is they let you determine the number of categories and subcategories in a very objective way.
Of course dendrograms are only as good as the data you put into them, and I don’t endorse basing taxonomy simply on genetic relatedness, but if I did, here’s how I’d interpret the above tree:
The first major split is between chimps & everyone else. This is consistent with two well recognized genera of hominins : Pan (i.e. chimps) and Homo (humans and near-humans).
Now within the Homo genus, we see another major split in the tree. Anatomically Modern Humans (AMH) vs Archaic Humans. Thus we can divide the homo genus into at least two major species.
Within the Archaic Humans we can further subdivide into major races: Denisovans and Neanderthals.
Now within our own species, AMH, the dendrogram shows three major races: Capoids, Congoids and Non-Africans.
I’m not saying I agree with this taxonomy since it was only based on genetic distance (much of which is junk DNA) but what’s great about using dendrograms is almost everyone looking at them will assign groups to the same categories and subcategories, even if they don’t use the same words (race, species, genus) to describe them. It’s wholly objective.
But what is needed is a dendrogram based on polygenic scores of actual phenotypes. That way people who have the same phenotypes caused by the same genomic architecture could be grouped together.
Unlike the above dendrogram, which groups based on how recently we share a common ancestor, we need to group based on how much of the common ancestor we share.
[update may 26, 2019: an earlier version of this article misspelled dendrogram]
[2nd update may 26, 2019: an earlier version of this article contained bragging that has since been removed]

Pumpkin, wouldn’t measuring things that are normally measure on achievement tests on IQ tests underestimate IQ, or are the secondary effects low enough not to really affect scoring that much?
Is this what Europeans used to look like?
https://www.theepochtimes.com/meet-the-ethiopian-boy-whose-rare-startling-blue-eyes-make-him-absolutely-unique_2936144.html
another comment peepee won’t approve:



no blue eyed european has eyes that color.
Could be photoshop. Epoch times? Lol
…so if Ethiopians were Europeans…or uh…Europeans were Ethiopians…
…and Ethiopians wuz the kangz who built the Pyramids…
…then…
…Europeans were the Egyptians who were the kangz who built the pyramids!
This is too confusing. I’ll just settle with taking ownership for what white people did, regardless. Like how Shakespeare was undeniably a black man, but because of our Eurocentric view of history, he’s considered white. Don’t you just hate that?
Why did you leave out sasquatchess?
good stuff. neat dendrogram.
James
From: Pumpkin Person
Sent: Saturday, May 25, 2019 11:06 PM
To: james.thompson@ucl.ac.uk
Subject: [New post] Genetic distance between humans, neanderthals & other primates
pumpkinpers
Thanks!
“Packing a 135 IQ, I was able to do what no blogger in behavioral genetics has ever done before: transform a genetic distance matrix into a dendogram, which looks at all the distances and creates the most parsimonious family tree:”
I suggest you rephrase this to sound less ego-maniacal. IQ denialists are going to read this and think you’re contemptible, and use that as the basis to dismiss everything you write. Use your 135 IQ to realise that.
okay fine, I removed it. But you can’t please everyone, so I usually don’t even try.
Ok, to be honest, linking everything you do to IQ is kind of egotistic (Pumpkin is not, he almost never does this). There are definitely good thinkers out there who have average IQs, and people who have good insight. Even below average IQs.
Pumpkin, how do you improve your social skills?
By watching other people with good social skills. You need a social skills role model, especially a male one. But if no such person has offered to coach you, then maybe watch documentaries about great politicians like bill Clinton & observe how they interact with the public.
The best advice is to just put yourself into social situations and learn from your mistakes.
But it’s a catch-22. You often can’t get the practice until you’re good in the first place.
Yea, people who are not good at baseball don’t “get the practice” untl they’re “good in the first place.”
Right, PP.
there’s no such thing as social skills. there’s just winning and not.
duh!
never be a phony. [redacted by pp, may 27, 2019]
you can’t FAKE being attractive or un-attractive. “social skills” is just an aesthetic. choose your own aesthetic and stick to it. if people don’t like it, it’s because they have bad taste.
This^ is an underrated comment.
Social skills is the capacity to adapt in society. But no there only one way to do it. And it’s often based on levels of individual compatibility with the society you live in.
Some people born in privileged places and with a privileged general biology. It’s easier for them to adapt in society they born.
Significant adaptability is when you born without any substantial extrinsic privilege but with a prodigious mind [specific privileged biology] in this basic set of skills.
Puppy goes to the shop:
Clerk: “Is there anything else you’d like sir?”
Puppy:”I would like recognition for creating a dendrogram on my blog please”.
Clerk: “I meant uh, something in store”
Puppy: “Yes, recognition for my dendrogram”
Clerk: “Ok, Im going to have to ask you to leave sir”
Puppy: “Thats not recognition for my dendrogram”.
Clerk: “Sir Im calling security”
Puppy: “Security is not recognition for my dendrogram”.
What percent of the variance in the comprehension subtest is attributed to social intelligence?
Pumpkin, would most bilinguals probably know the vocabulary on the easy items of the similarities subtest- or would they most likely know all of the, since they are easy words.
PP, in one of your comments in the previous article you said that most people would agree with tulsi gabbard on foreign policy. I think they will …if they believe here lie implying U.S and Russia will nuke each other over iran.
I am posting this comment here because I am not able to find that comment and also because all threads here are open threads.
I meant if they believe her lie.
why won’t you approve my billy herrington videos and the denzel doll i made?
I don’t approve textless comments
do you find this hilarious?
1. yes.
2. you have autism.
maradona has got to be 90% native american and he may be the GOAT for his sport. up there with pele and zidane and muller etc.
no native american athlete has achieved as much? right?
the point is that south asian men are effeminate and suck at all sports. they are the world’s worst athletes.
why is imran khan the closest thing to a steph curry mucher?
Thats interesting but the goat is probably messi. But i dont know i never saw maradonnna pele or di stefano in their primes.
meh it doesn’t matter if they suck at sports.
https://en.m.wikipedia.org/wiki/List_of_Indian_inventions_and_discoveries
notice peepee identifies capoids as a major race but does not identify south indians/abos/papuans as a major race even though they outnumber capoids 1,000 to 1 at least.
the bantus took no prisoners.
Did you just include south Indians in the abo category?
https://www.tomsguide.com/us/soft-photopolymer-cuboid-computing,news-30158.html
Not to mention a lot of stuff in the earlier list I posted is from south Indians. Also, the person called Arya bhatta mentioned in that list was darkskinned.
On another note: Arya/ Aryan is not an ethnicity it was a title and also sometimes used as a persons name.
yes. south indians are closely related to australian abos. see the picture in peepee’s dendropenis and see orson welles’s The Journey of Man.
street shitters never invented shit.
https://en.wikipedia.org/wiki/The_Journey_of_Man
Define closely related. Also what would you say about the list I posted?
And the other link?
also notice peepee identifies all non-blacks as “north africans”.
this is because peepee is black.
Non-African not north-African
peepee is hilarious…
UN-intentionally.
From a genomic perspective there are just 3 races: blacks, blacks & non-blacks
Humans and mice share 99% of dna. Erg9 dna is irrelevant in measuing differences. Unless you scale it or something.
No it totally matters that’s why I’m half mouse half human.
Stop being xenophobic against people of rodent-descent please….
Pumpkin, do you approve of comments yourself?
Puppy hires a black person tru affirmitve action to read and mod all comments. This is why the pro africa slant on the blog is strong.
it’s because peepee is black.
If I took a DNA test it would likely show 0% sub-Saharan ancestry. You on the other hand would likely have a small percentage just from the slaves your ancestors probably raped
Yes, this place, oddly enough, has “a lot of” black of people compared to the other blogs I’ve lurked in. And Robert Lindsay also had a hefty sum of blacks. And both Robert and Pumpkin are liberals. I don’t know where I’m going with this.
Quite surprised about Lindsay’s black population.
Billy,
Its because of 3 reasons.
In both PP´s and Lindsays blog there has been a period of “egi arguments” where different groups of people jab at eachoder, or people discuss other groups in a holistic sociological manner, but with an overwhelming amount of edgy humor. The only way to keep the good aspects of cordinal discussion together with the engagement that comes with edgy humor is to have a moderator that makes sure that the website doesnt get overcrowded with outwardly nazi troglodite beliefs (since the internet and hbd followers are filled with whites) that would make all other people feel like their words dont matter, yet try to maintain a diversity of views by making the more established ideologies raise their standard in intelectuall contribution and be nicer (pp dungeon). Thusly you get a bunch of non-whites to jump out of the closet and the higher iq ones stay becuase theres no other place to debate without getting berated by a lack of engagement from the community.
Why PP and Lindsay are the only ones who have done this successfully is becuase they are high iq/high cultured enough to know how to make posts that higher iq people will enjoy and how to meticolously moderate the more vulgar elements of the hbd community. The lack of conformity and amound of higher iq readers with various ideas will intice the higher iq nazis like former poster Jimmy to argue since theres more to gain in writing in those two platforms than, say, the unz review, becuase of the audience that the moderators form.
Its a shame that both Robert and PP banned their crazy elements. I miss Jimmy.
The person mentioned in the article kalaichelvi saravanamuttu is south indian.
The most annoying thing is when geneticists in the media talk to dumb dumbs and say that human beings share 99.9% of DNA so theres no such thing as race and they NEVER mention that mice share 99% or chimps. So most of DNA is junk and they know it. Geneticists have known about race since the late 19th century when Darwins cousin kept talking about it and in the past 50 years the danes have successfuly cowed anyone with brains out of the profession and replaced them with autistic idiots that parrot the party line that is completely fake news.
No it has to do with chromosomes and certain parts of junk DNA regulating the coding genes, so much of “junk DNA” is really relevant. It wouldntake sense to have chimps be 99% similar to us, becuase that would mean that chimps would come out of human females.
Most DNA really is junk
Ok most is but we are not sure by how much and the 98.8 statistic often named is suspect and too brief in context to explain how many genes have causial effects on our phenotypic diversity.
[redacted by pp, may 27, 2019] those who are aware and try to hide it usually can’t. even if they say nothing overtly anti-danish they will be punished unless they successfully pretend to be pro-danish. everyone is afraid to be the first person to stop clapping for denmark.
i wouldn’t be surprised if people with german surnames or storm trooper looks are much less prominent in modern america than their numbers and ability would merit because danish hatred of germans.
eisenhower and bush the only german surname presidents despite germans being the largest slice of the white immigrant pie.
as you can see: germans dominate.

those who are aware and try to hide it usually can’t.
This statement shows your severe autism. So severe it’s probably visible from a mile away because you likely rock while you walk.
The only reason someone as autistic as you knows about the Danes is because you’re a high IQ racist white male. If your IQ were normal or your race black or Asian, you’d be as clueless on this topic as you are on other social issues
Its amazing how the amount of scientists in the west has probably quintupled or so since college was made more available in the 50s (unfortunately to danes), and the amount of scientific knowledge gained actually seen a remarkable slowdown.
I think I read somewhere that the only useful thing that got invented in STEM since the 60s was modern statistics.
It’s a very interesting phenomenon. Maybe it also has to do with the fact that the brightest students are more likely to chose majors like business and poli sci than they were 60+ years ago.
At university, my general elective science professors were consistently the LEAST intelligent ones.
steve shoe made the same claim. he attributes it to selection for “hoop jumpers” and thus de-selection for original thinkers. it doesn’t occur to him that he is a hoop jumper because no self-awareness because chinaman.
STEM includes computer crap pill. yuge developments since the 60s.
HAART was developed with billions. now you can have anal sex all day long and not die from AIDS. what a good use of resources.
statins are the biggest medical breakthrough if they actually do prevent atherosclerosis.
SSRIs are better than the tricyclics of the 60s.
but just as in the 60s, if you get cancer past the age of 50 you’re usually screwed. or so i’ve been told. the progress in cancer is almost entirely in juvenile and young adult cancers which are very rare.
Dane puppets in the media and useful idiots in general lib media are annoying. But the really annoying ones are actual danes lying and lying about syria and iran and russia. It is nauseating to watch it.
what are your thoughts on this conspiracy theory:
The deal with India paying rupees to Iran is by Iran to make India buy oil from it. And India has been friendly with Iran to counter Pakistan and also to prevent Iran to fall too much into China’s arms.
Puppys hero Oprah is a uesful idiot for the danes. She might not agree on foreign policy but all the social stuff is up her alley. I think she even called for reparations once. Thats the type of magic negro were dealing with,.
No, Oprah has not called for reparations and even if she had, I fail to see how that would make her a useful idiot. In fact you could argue that the smartest thing blacks have ever done is align themselves with the danes since the danes were instrumental in getting blacks the civil rights, affirmative action & media exposure they lacked under WASP rule. There’s a difference between being useful and being a useful idiot. The latter sacrifices their own interests to help the elite while the former has a mutually beneficial relationship. Oprah has advanced both her financial interests by making billions and her genetic interests by help creating a much more pro-black society than existed when she started, so she’s nobody’s fool.
“Oprah has advanced .. her genetic interests by help creating a much more pro-black society”
Black genetic interests in action in our more pro-black society:
34 shot, 5 fatally, so far in Chicago during Memorial Day weekend
Black life expectancy has increased since oprah hit the scene:
https://www.washingtonpost.com/news/to-your-health/wp/2017/05/02/cdc-life-expectancy-up-for-blacks-and-the-racial-gap-is-closing/?noredirect=on&utm_term=.e5b26b700fb7
Interestingly black life expectancy has increased in Brazil, without Oprah influence.
But in the u.s. the racial life span gap declined. Not saying oprah deserves the credit but I am saying that elite policy is not anti-black in the u.s.
What is a black person in Brazil?
Elite policy is pro black because of danes, not oprah.
See you’re contradicting yourself because your IQ’s not that high
First you say she’s useful idiot for Danes
Then you say Danes advance black interests
Well if they’re advancing her group’s interests, there’s nothing idiotic about her being useful to them (assuming she even is)
Oprah is a dancing bear. The type you see at the circus. Just because the circus hires bears doesnt mean the circus is pro bear.
but you’ve been telling us for years that danes have privileged bears in our society, to the point where being a bear is like winning a lottery.
and your metaphor is ironic, because Oprah was virtually the first black icon to literally not do stuff like dancing. Unlike other black icons, she got rich and famous interviewing and discussing issues, not dancing, singing or throwing basketballs.
”What is a black person in Brazil?”
Just like Oprah, but poor.
So you call me stupid abd then moderate my explanation. Great.
fine, i’ll unmoderate it
First time my whole life i saw an east asian in a blue collar job (in the west) today. Funny.
Black women ads all over puppys blog as usual. Google must have picked up the continuous use of rhe word oprah here.
Loaded,
How do you think Africans stack up to Chinese intellectually, economically and military wise.
I dunno, but I think the Chinese are rude and entitled and might be psychic. I guess they’re like an alien race on Earth that I find to be very outrageous. I see race almost like species so it’s hard to see what’s actually happening when there are so many subdivisions of people.
The Chinese may lack serious morals, but I’m not sure about psychic.
Yeah maybe psychopathic.
actually “bush” is an english name. i mis-remembered as usual.
so the SOLE president with a german surname, prior to trump, was eisenhower?
there was also “hoover”, an anglicized version of “huber”.
so only 2 presidents.
“trump” is an anglicized version of german “drumpf”.
van buren and roosevelt were ny dutch.
Just because presidents dont have a german surname doesnt mean they dont have german ancestry. And i think bush was originally ‘busch’.
top 50 include: anschutz, mars?, hamm, schmidt, ergen, frist
blacks are worse off today than they were in the 56s
No they’re not you racist rapist. Only economically. The bw life span gap has decreased & blacks now have more status.
american negroes are much more anti-jew than american whites…in the open.
I know that mug of pee. I’m just pointing out the inconsistency of you & pill
On the one hand you say blacks are privileged under Jewish rule & whites are oppressed, on the other hand you say blacks are dumb puppets to side with Jewish elites
How is it dumb to side with an elite that is privileging your group?
Or do you admit it’s actually whites who are privileged?
You can’t have your cake & eat it too.
If segregation existed for about 100 years black IQ would be at 90. We had a good thing going, but people had to ruin it.
https://twitter.com/califuneral/status/1131255026523840513
”ayrians”
Pumpkin, if you have a series of 4 trials of 7 letter letter span, to someone continuously- would they show improvement? Will the scores be the same pretty much, the letters are all in a different pattern- and they have different letters too.
Also, I believe that the practice effect for symbol search and digit span has some aspect of grasping something for a second time. You are looking and hearing the patterns for a second time.